See the GitHub repository here.
I took a Bioinformatics module which explores the development of methods and software tools for understanding biological data. I was tasked with producing a tool that searches a protein sequence for any matched motifs, providing the position of each motif found. In particular, the tool had to detect regions of protein sequences which could form amyloid associations; these have been associated with neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease.
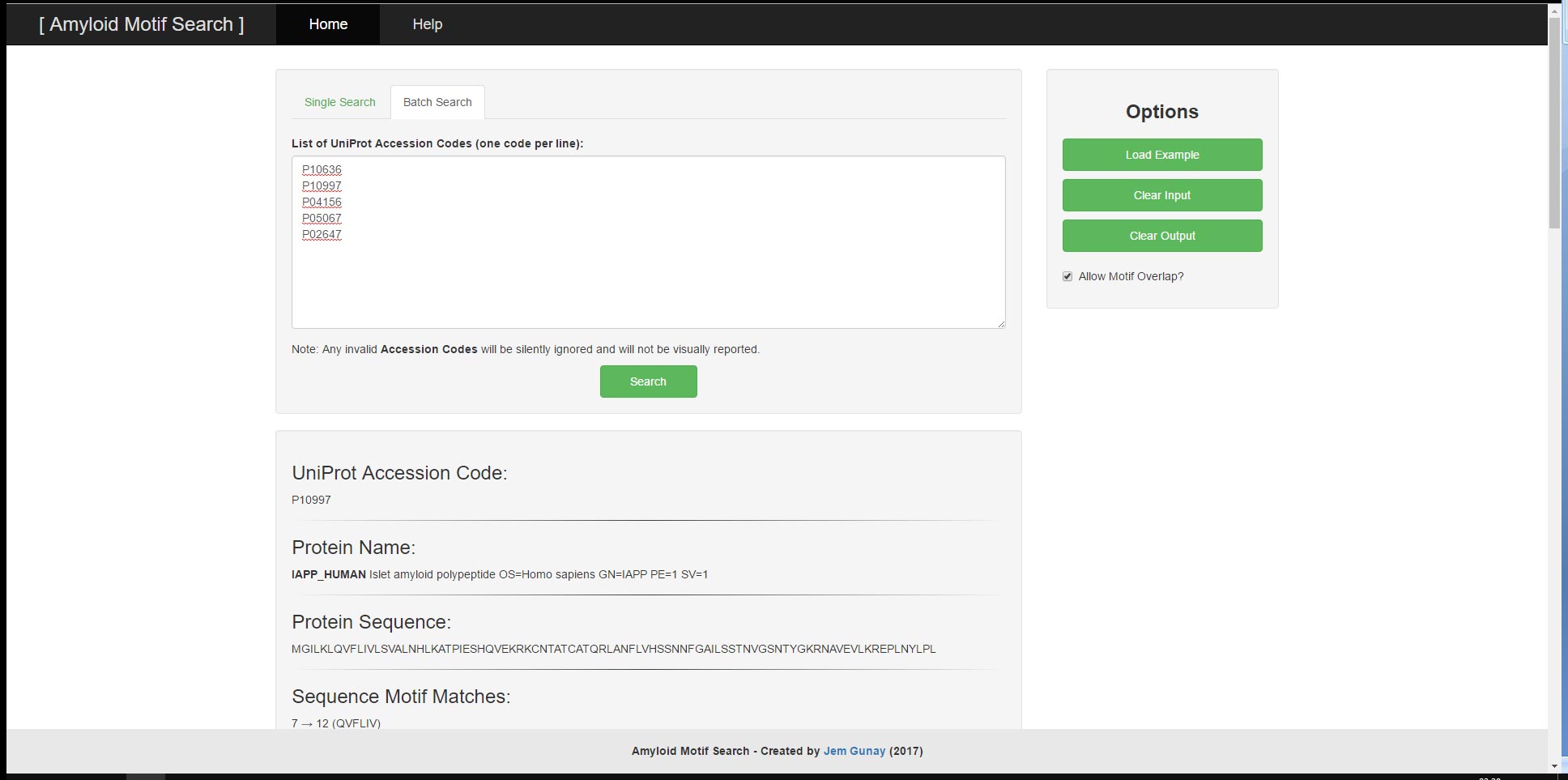
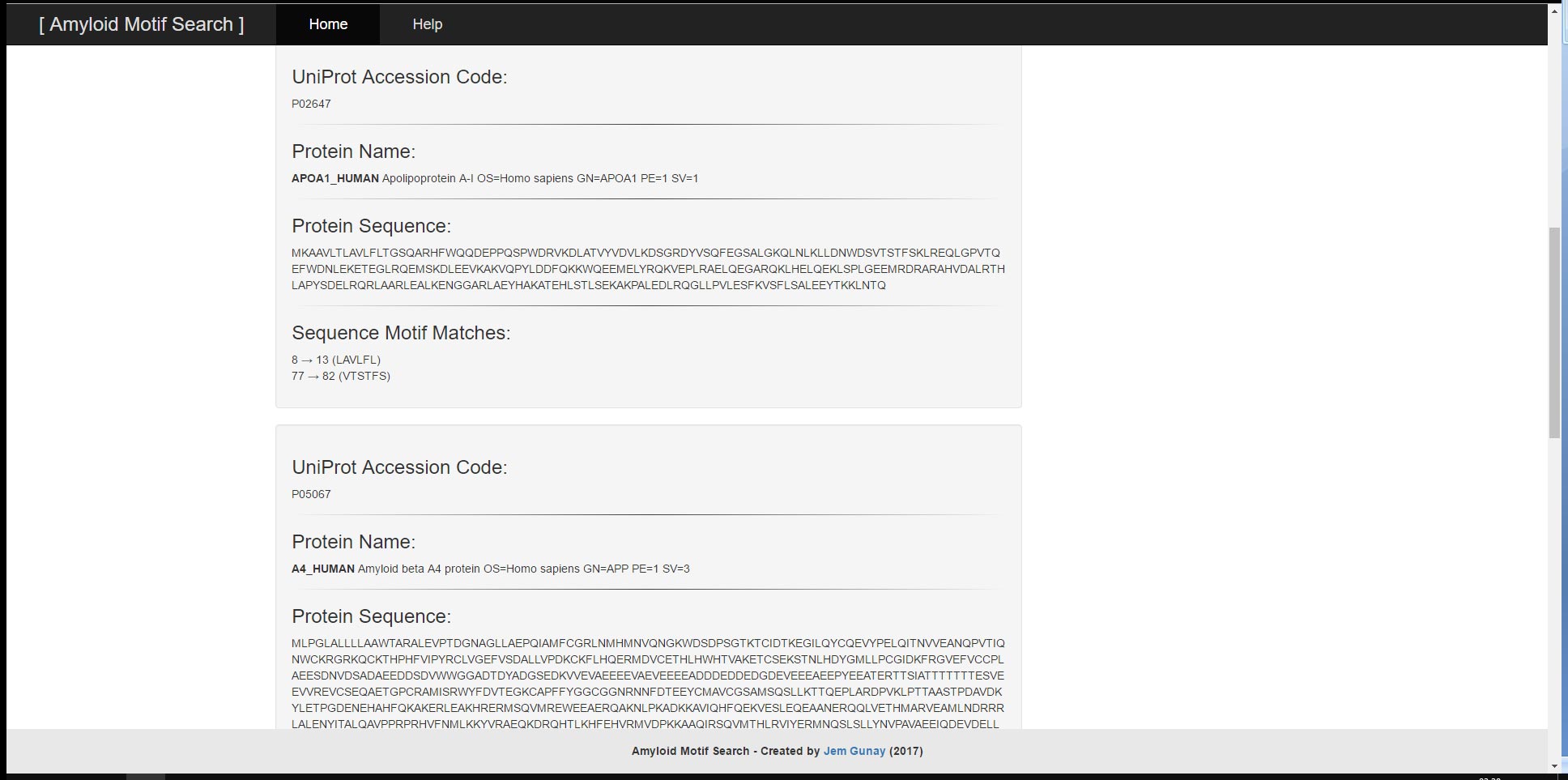
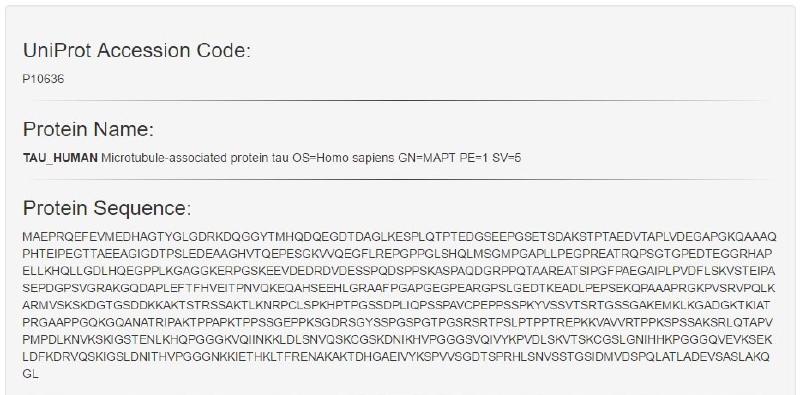
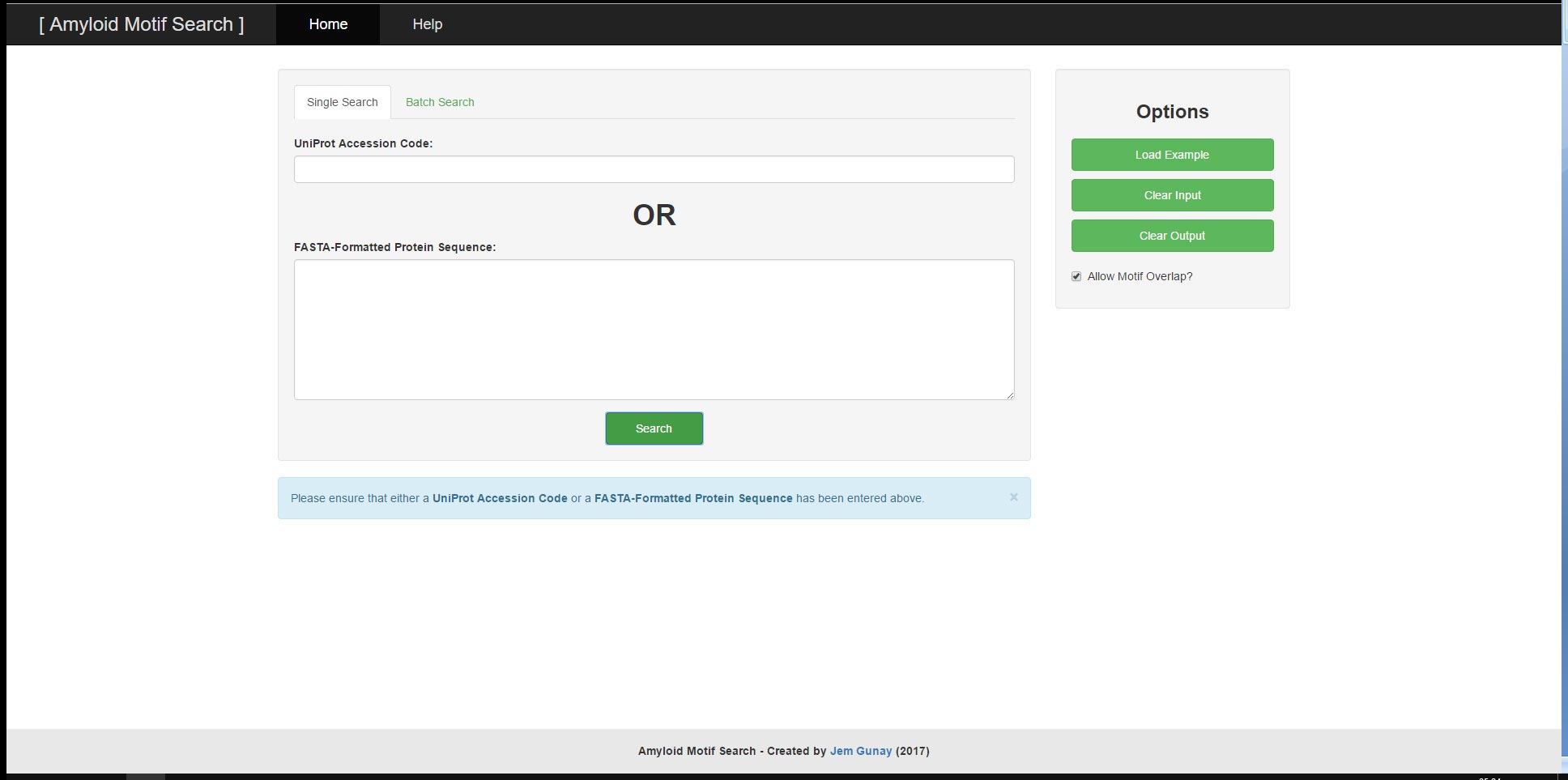
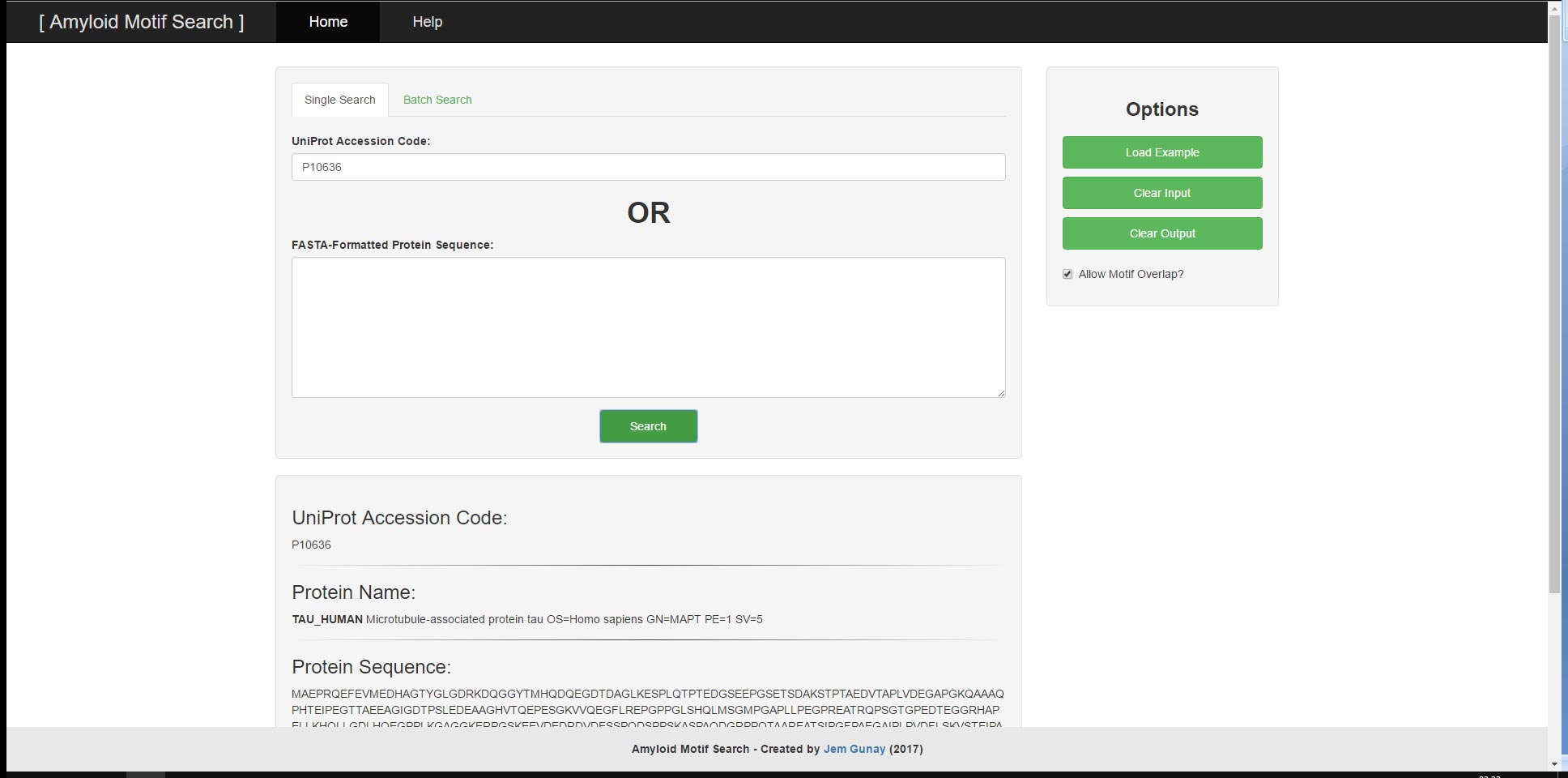
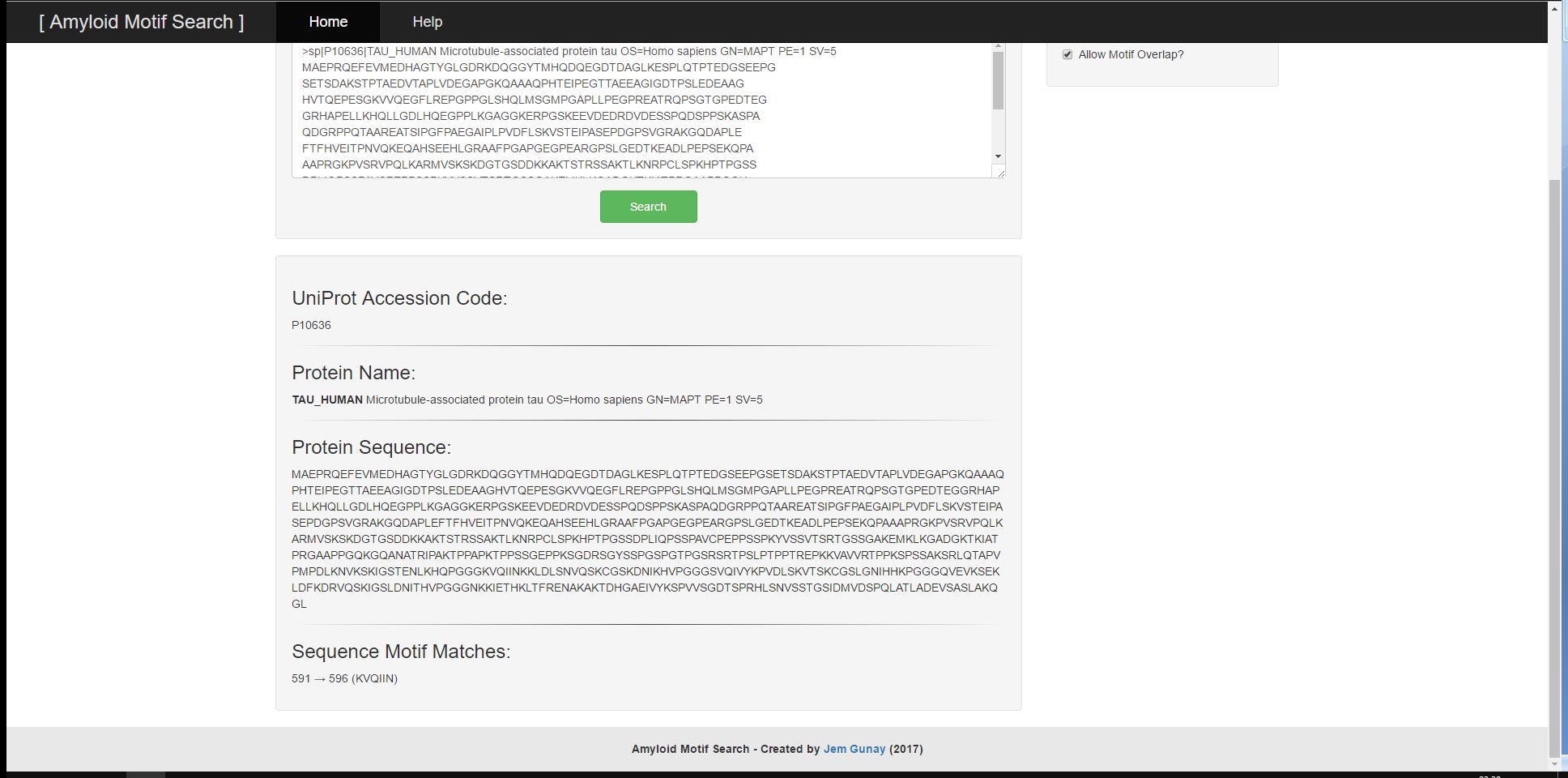
User provided input can consist of either a UniProt Accessor Code or a FASTA-Formatted Protein Sequence and will result in the output of the UniProt Accessor Code, the FASTA-Formatted Protein Sequence, the Protein Name and the corresponding Sequence Motif Matches. The tool has two main functions; the ability to search a singular protein sequence for patterns and the ability to search through a batch of protein sequences by their UniProt Accessor Codes all at once. For both functions, there are example inputs and options to clear input and output. The search algorithm and input validations utilise regular expressions to search for relevant motifs and to prevent malformed user input from leading to undesirable results or performance.
The tool was developed using Python 2.7, as well as HTML5, CSS3 and JavaScript. The Bootstrap front-end framework was used to design the front-end of the system efficiently and the jQuery library was used to write more elegant dynamic code. It can run offline on local web servers which allow the execution of Python files via CGI (Common Gateway Interface) assuming sufficient permissions have been set.